Databricks Certified Associate Developer for Apache Spark 3.5 - Python Databricks Certified Associate Developer for Apache Spark 3.5 Exam Practice Test
A data engineer is asked to build an ingestion pipeline for a set of Parquet files delivered by an upstream team on a nightly basis. The data is stored in a directory structure with a base path of "/path/events/data". The upstream team drops daily data into the underlying subdirectories following the convention year/month/day.
A few examples of the directory structure are:
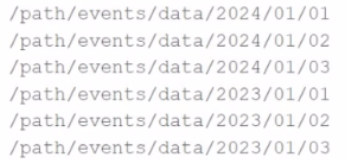
Which of the following code snippets will read all the data within the directory structure?
Answer : B
To read all files recursively within a nested directory structure, Spark requires the recursiveFileLookup option to be explicitly enabled. According to Databricks official documentation, when dealing with deeply nested Parquet files in a directory tree (as shown in this example), you should set:
df = spark.read.option('recursiveFileLookup', 'true').parquet('/path/events/data/')
This ensures that Spark searches through all subdirectories under /path/events/data/ and reads any Parquet files it finds, regardless of the folder depth.
Option A is incorrect because while it includes an option, inferSchema is irrelevant here and does not enable recursive file reading.
Option C is incorrect because wildcards may not reliably match deep nested structures beyond one directory level.
Option D is incorrect because it will only read files directly within /path/events/data/ and not subdirectories like /2023/01/01.
Databricks documentation reference:
'To read files recursively from nested folders, set the recursiveFileLookup option to true. This is useful when data is organized in hierarchical folder structures' --- Databricks documentation on Parquet files ingestion and options.
10 of 55.
What is the benefit of using Pandas API on Spark for data transformations?
Answer : A
Pandas API on Spark provides a distributed implementation of the Pandas DataFrame API on top of Apache Spark.
Advantages:
Executes transformations in parallel across all nodes and cores in the cluster.
Maintains Pandas-like syntax, making it easy for Python users to transition.
Enables scaling of existing Pandas code to handle large datasets without memory limits.
Therefore, it combines Pandas usability with Spark's distributed power, offering both speed and scalability.
Why the other options are incorrect:
B: While it uses Python, that's not its main advantage.
C: It runs distributed across the cluster, not on a single node.
D: Pandas API on Spark uses lazy evaluation, not eager computation.
PySpark Pandas API Overview --- advantages of distributed execution.
Databricks Exam Guide (June 2025): Section ''Using Pandas API on Apache Spark'' --- explains the benefits of Pandas API integration for scalable transformations.
===========
A developer is working with a pandas DataFrame containing user behavior data from a web application. Which approach should be used for executing a groupBy operation in parallel across all workers in Apache Spark 3.5?
A)
Use the applylnPandas API
B)

C)


Answer : A
The correct approach to perform a parallelized groupBy operation across Spark worker nodes using Pandas API is via applyInPandas. This function enables grouped map operations using Pandas logic in a distributed Spark environment. It applies a user-defined function to each group of data represented as a Pandas DataFrame.
As per the Databricks documentation:
'applyInPandas() allows for vectorized operations on grouped data in Spark. It applies a user-defined function to each group of a DataFrame and outputs a new DataFrame. This is the recommended approach for using Pandas logic across grouped data with parallel execution.'
Option A is correct and achieves this parallel execution.
Option B (mapInPandas) applies to the entire DataFrame, not grouped operations.
Option C uses built-in aggregation functions, which are efficient but not customizable with Pandas logic.
Option D creates a scalar Pandas UDF which does not perform a group-wise transformation.
Therefore, to run a groupBy with parallel Pandas logic on Spark workers, Option A using applyInPandas is the only correct answer.
A data engineer wants to write a Spark job that creates a new managed table. If the table already exists, the job should fail and not modify anything.
Which save mode and method should be used?
Answer : A
The method saveAsTable() creates a new table and optionally fails if the table exists.
From Spark documentation:
'The mode 'ErrorIfExists' (default) will throw an error if the table already exists.'
Thus:
Option A is correct.
Option B (Overwrite) would overwrite existing data --- not acceptable here.
Option C and D use save(), which doesn't create a managed table with metadata in the metastore.
Final Answer: A
A data engineer wants to create a Streaming DataFrame that reads from a Kafka topic called feed.
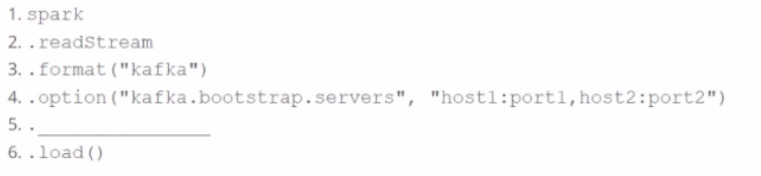
Which code fragment should be inserted in line 5 to meet the requirement?
Code context:
spark \
.readStream \
.format("kafka") \
.option("kafka.bootstrap.servers", "host1:port1,host2:port2") \
.[LINE 5] \
.load()
Options:
Answer : A
To read from a specific Kafka topic using Structured Streaming, the correct syntax is:
python
CopyEdit
.option('subscribe', 'feed')
This is explicitly defined in the Spark documentation:
'subscribe -- The Kafka topic to subscribe to. Only one topic can be specified for this option.' (Source: Apache Spark Structured Streaming + Kafka Integration Guide)
'subscribe -- The Kafka topic to subscribe to. Only one topic can be specified for this option.' (Source: Apache Spark Structured Streaming + Kafka Integration Guide)
B . 'subscribe.topic' is invalid.
C . 'kafka.topic' is not a recognized option.
D . 'topic' is not valid for Kafka source in Spark.
A Spark application is experiencing performance issues in client mode because the driver is resource-constrained.
How should this issue be resolved?
Answer : C
In Spark's client mode, the driver runs on the local machine that submitted the job. If that machine is resource-constrained (e.g., low memory), performance degrades.
From the Spark documentation:
'In cluster mode, the driver runs inside the cluster, benefiting from cluster resources and scalability.'
Option A is incorrect --- executors do not help the driver directly.
Option B might help short-term but does not scale.
Option C is correct --- switching to cluster mode moves the driver to the cluster.
Option D (local mode) is for development/testing, not production.
Final Answer: C
19 of 55. A Spark developer wants to improve the performance of an existing PySpark UDF that runs a hash function not available in the standard Spark functions library.
The existing UDF code is:
import hashlib
from pyspark.sql.types import StringType
def shake_256(raw):
return hashlib.shake_256(raw.encode()).hexdigest(20)
shake_256_udf = udf(shake_256, StringType())
The developer replaces this UDF with a Pandas UDF for better performance:
@pandas_udf(StringType())
def shake_256(raw: str) -> str:
return hashlib.shake_256(raw.encode()).hexdigest(20)
However, the developer receives this error:
TypeError: Unsupported signature: (raw: str) -> str
What should the signature of the shake_256() function be changed to in order to fix this error?
A.
def shake_256(raw: str) -> str:
B.
def shake_256(raw: [pd.Series]) -> pd.Series:
C.
def shake_256(raw: pd.Series) -> pd.Series:
D.
def shake_256(raw: [str]) -> [str]:
Answer : C
Pandas UDFs (vectorized UDFs) process entire Pandas Series objects, not scalar values. Each invocation operates on a column (Series) rather than a single value.
Correct syntax:
@pandas_udf(StringType())
def shake_256(raw: pd.Series) -> pd.Series:
return raw.apply(lambda x: hashlib.shake_256(x.encode()).hexdigest(20))
This allows Spark to apply the function in a vectorized way, improving performance significantly over traditional Python UDFs.
Why the other options are incorrect:
A/D: These define scalar functions --- not compatible with Pandas UDFs.
B: Uses an invalid type hint [pd.Series] (not a valid Python type annotation).
PySpark Pandas API --- @pandas_udf decorator and function signatures.
Databricks Exam Guide (June 2025): Section ''Using Pandas API on Apache Spark'' --- creating and invoking Pandas UDFs.